All Technologies Used
Motivation
The client approached Azati to reduce the time and effort required for multi-sequence analysis, improve the accuracy and relevance of search results, and automate the comparison of multiple nucleotide or protein sequences across large biological databases. The goal was to streamline research workflows, minimize manual errors, and enable scientists to efficiently identify complex genetic patterns and alignments.
Main Challenges
Biological scientists needed a tool to search for multiple nucleotide or protein sequences at once, which was critical for identifying CDRs, chimeric constructs, and recombinant plasmids. At the time, no such tool existed in the market. Azati proposed the design and development of a new multiple sequence search feature from scratch.
Researchers lacked the ability to correlate multiple sequences across various claims within the same patent document, limiting the accuracy and depth of results. Azati addressed this by building an advanced scoring system and enhanced interface to ensure precise multi-alignment tracking.
Our Approach
Want a similar solution?
Just tell us about your project and we'll get back to you with a free consultation.
Schedule a callSolution
Multi-Query Input
- Input multiple nucleotide or peptide sequences at once
- Perform simultaneous searches across multiple biological databases
- Streamline research workflows with batch query support
- Reduce repetitive manual searches
Advanced Document Scoring
- Prioritize documents with the highest number of matching sequences
- Highlight key alignment regions for easy identification
- Ensure relevance of search results through advanced scoring
- Support identification of overlapping or related sequences in patents
GPU-Accelerated Alignment
- Leverage GPU processing for high-speed alignment
- Maintain high accuracy in sequence comparisons
- Handle large datasets efficiently
- Accelerate research workflows by reducing computational time
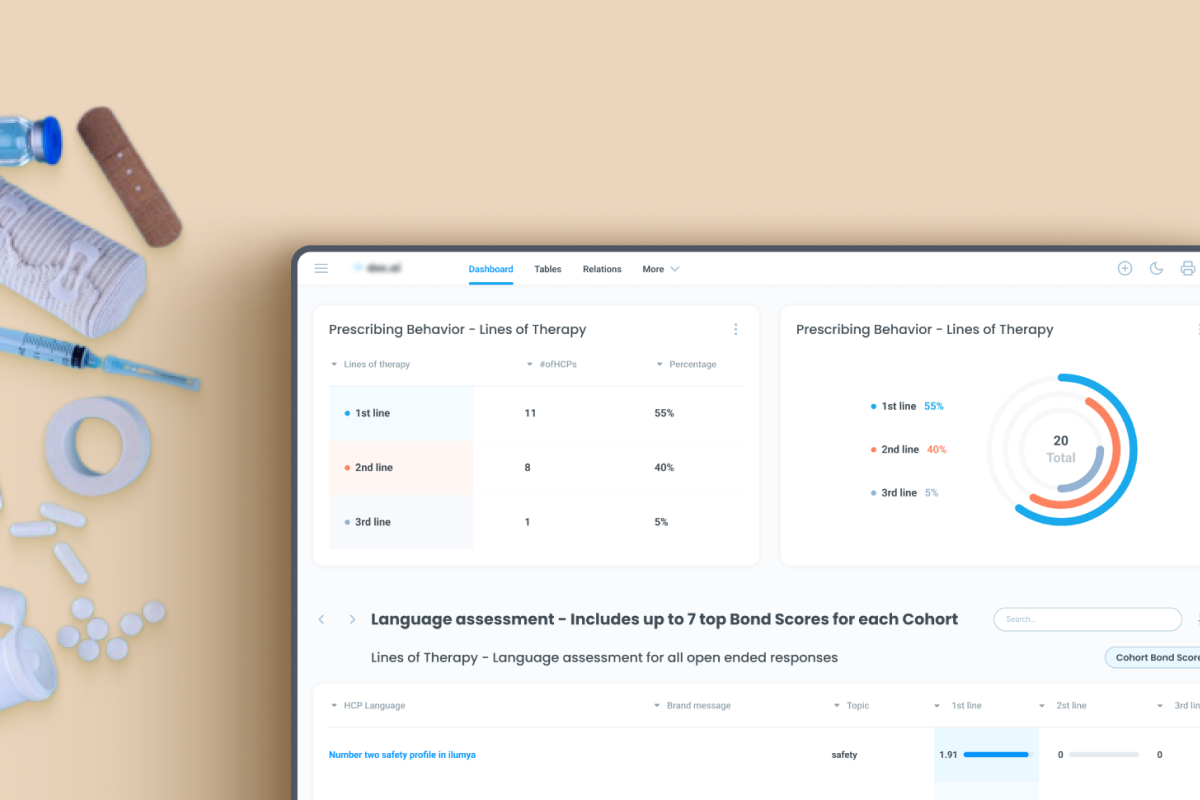
Multi-Sequence Patent Analysis
- Search for multiple sequences within the same patent document
- Identify sequences present in different claims
- Support advanced IP and research analysis
- Visualize sequence overlaps for easier interpretation
Exportable Reports
- Export results in multiple file formats
- Include visual sequence alignments in reports
- Enable easy sharing with research teams
- Integrate results into downstream bioinformatics pipelines
User-Friendly Interface
- Visualize sequence alignments clearly
- Switch between individual and combined alignment views
- Navigate large datasets efficiently
- Simplify interpretation of complex multi-sequence search results
Business Value
Accelerated Research: Enabled researchers to perform multi-sequence queries significantly faster, reducing project timelines and increasing the speed of genetic discovery.
Increased Accuracy: Advanced scoring and GPU-accelerated alignments improved precision in identifying relevant genetic sequences and patent claims.
Enhanced Workflow: Exportable reports and a user-friendly interface simplified data interpretation and collaboration, streamlining daily research activities.
Improved Patent Analysis: Multi-sequence patent correlation allowed scientists to identify overlapping sequences and track complex genetic constructs across claims, improving IP research and compliance.
Time and Resource Efficiency: High-performance GPU processing reduced computational overhead, allowing teams to focus on analysis rather than waiting for results.